Bioinformatics
Sphetcher: new method for sketching large single-cell datasets
03.05.2020
The massive size of single-cell RNA sequencing datasets often exceeds the capability of current computational analysis methods to solve routine tasks such as detection of cell types. In collaboration with Khaled Elbassioni (Khalifa University of Science and Technology, UAE), Van Hoan Do from the Canzar lab has developed method Sphetcher that efficiently picks representative cells that accurately capture the geometry of the transcriptional space occupied by the original data.
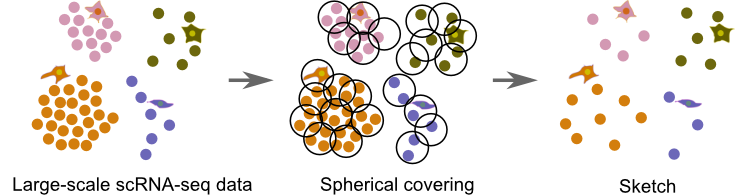
For a large single-cell RNA sequencing dataset (left), Sphetcher uses a disk-friendly optimization algorithm to compute a smallest size set of spheres of a fixed radius that cover all cells (middle). One representative cell (the center) from each sphere is selected into the final spherical sketch (right)
The resulting sketch of single cells highlights rare cell types, facilitates visualization and sharing of large datasets and accelerates downstream analyses such as trajectory inference. In addition, Sphetcher’s underlying optimization scheme allows to include fairness aspects that can encode prior biological or experimental knowledge. Their experiments show that a fair sampling of single cells can inform the inference of the trajectory of human skeletal muscle myoblast differentiation.

