Cell Biology
Your number’s up!
17.04.2020
mRNAs program the synthesis of proteins in cells, and their functional lifetimes are dynamically regulated. Roland Beckmann and his team have now shown why blueprints that are more difficult to decipher have shorter lifetimes than others.
The control of gene expression is a fundamental component of living systems. The term refers to the suite of mechanisms that determines how the hereditary information encoded in the DNA genome of every cell is selectively transcribed into messenger RNAs (mRNAs), and then translated by ribosomes into proteins. Because the set of proteins synthesized in a cell defines its structure and biochemical capacities, every step in the process must be tightly regulated. One of the modules of this regulatory system is dedicated to the timely destruction of mRNAs in response to changing conditions.
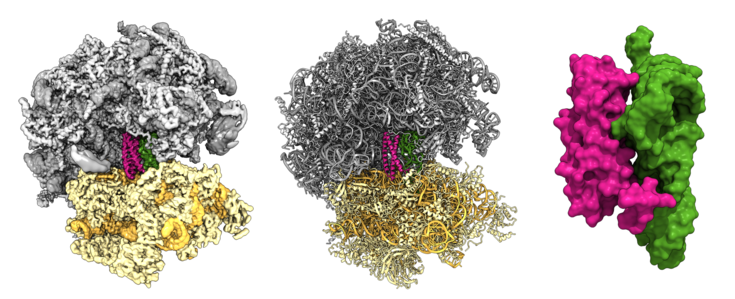
Structure of the Not5-ribosome complex. Cryo-EM density of the Not5-80S ribosome complex (left). Molecular model of the Not5-80S ribosome complex (middle). Surface representation of the Not5-tRNA interaction (right). The 40S ribosomal subunit is colored in beige, the 60S subunit is colored in grey, the tRNA is colored in green and Not5 is colored in pink. Source: R. Buschauer
An international team led by Professor Roland Beckmann, in collaboration with Jeff Coller (Case Western Reserve University, Cleveland, USA) and Toshifumi Inada (Tohoku University, Sendai, Japan) has now worked out the detailed structure of a protein complex that is involved in mRNA degradation, and dissected its mode of action. The results of the new study, which appears in the leading journal Science, explain how and why the lifetime of an mRNA molecule is linked to the rate of synthesis of the protein it encodes.
“Statistical data had already revealed that the lifetime of an mRNA is correlated with speed of the ribosome during synthesis of its protein product,” says Robert Buschauer, a PhD student in Beckmann’s group and lead author of the new paper. “But the molecular basis for this relationship was completely unknown.
More information please visit LMU.de/news
Original Publication:
The Ccr4-Not complex monitors the translating ribosome for codon optimality
Buschauer R, Matsuo Y, Sugiyama T, Ying-Hsin Chen Y, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y,
Nobuta R, Gilmozzi A, Berninghausen O, Tesina P, Becker T, Coller J, Inada T, Beckmann R.
Science, 142–149 (2020). https://doi.org/10.1126/science.aay6912

