Structural Biology
Structural basis of non-canonical nucleosome remodeling
29.06.2023
Loss of H2A-H2B histone dimers is a hallmark of actively transcribed genes, but how the cellular machinery functions in the context of non-canonical nucleosomal particles remains largely elusive. The Eustermann group in collaboration with the Hopfner lab report the structural mechanism for ATP-dependent chromatin remodeling of hexasomes by the INO80 complex. They show how INO80 recognizes non-canonical DNA and histone features of hexasomes emerging from the loss of H2A-H2B. A large structural re-arrangement switches the catalytic core of INO80 into a distinct, spin-rotated mode of remodeling, while its nuclear actin module remains tethered to long stretches of unwrapped linker DNA. Direct sensing of an exposed H3-H4 histone interface activates INO80, independently of the H2A-H2B acidic patch. Their findings reveal how the loss of H2A-H2B grants remodelers access to a different, yet unexplored, layer of energy-driven chromatin regulation.
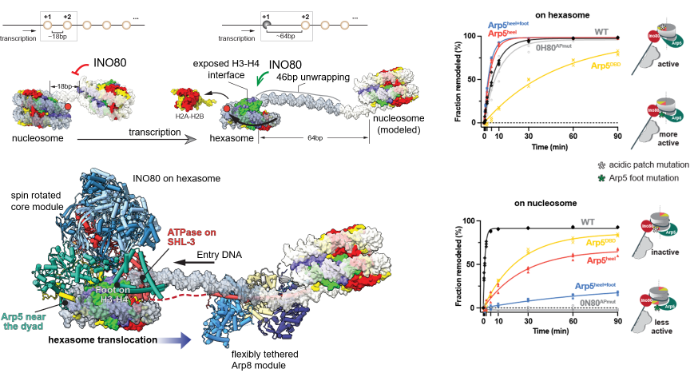
Structural model of non-canonical nucleosome remodeling at sites of transcription (source: Hopfner lab)
Original publication:
Hexasome-INO80 complex reveals structural basis of non-canonical nucleosome remodeling
Zhang M, Jungblut A, Kunert F, Hauptmann L, Hoffmann T, Kolesnikova O, Metzner F, Moldt M, Weis F, DiMaio F, Hopfner KP, Eustermann S
Science 2023

