Structural Biology
The art of unpacking
11.04.2018
DNA is tightly packed into the nucleus of a cell. Nevertheless, the cellular machinery needs to constantly access the genomic information. Karl-Peter Hopfner and his team now reveal the inner workings of a molecular motor made of proteins which packs and unpacks DNA.
The genomic DNA of higher organisms is compacted in a highly condensed form known as chromatin. The DNA is tightly wound around a myriad of tiny histone spools called nucleosomes. A single human cell, for instance, accommodates in this manner about two meters of DNA. However, genes must be constantly transcribed into messenger RNAs to direct protein synthesis. Moreover, the entire DNA must be replicated before cell division and DNA damage needs to be repaired. Thus, there must be way to actively grant access to the genome.
This is when chromatin remodelers come into play. Chromatin remodelers have an essential role as they are molecular machines: they unpick and unpack segments of the DNA by sliding nucleosome spools back and forth, replacing individual histones, freeing up the DNA for transcription, and finally compacting it again, when the job is done. Since all of this happens in a highly dynamic fashion, chromatin remodelers enable cells to react rapidly to alterations in their environment.
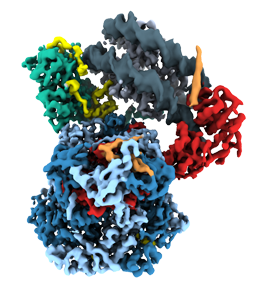
The architecture of INO80's nucleosome remodeling core.
So far, however, very little is known about what remodeling proteins look like and how they go about doing what they do. In molecular terms, functional remodelers are often very large complexes comprising many different protein components, whose coordinated action makes them akin to molecular machines. These features also make it very difficult to determine their detailed structure. Karl-Peter Hopfner and his team has now used cryo-electron microscopy to reconstruct the three-dimensional structure of the nucleosome-sliding remodeler INO80 (which itself consists of 15 subunits) bound to a single nucleosome. The structure of the complex revealed in the new study sheds new light on the function and mode of action of chromatin remodelers in general. The findings appear in the leading journal Nature.
More information please visit LMU.de/news
Original Publication:
Structural basis for ATP dependent chromatin remodelling by the INO80 complex.
Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner KP.
Nature. 2018 April 11; doi:10.1038/s41586-018-0029-y. PubMed

