Bioinformatics
TACCO: new framework for analysis of single-cell and spatial omics data
16.02.2023
Simon Mages and the Klughammer lab in collaboration with the Regev and Nitzan labs developed TACCO, a new framework for the efficient analysis of single-cell and spatial omics data (Transfer of Annotations to Cells and their COmbinations).
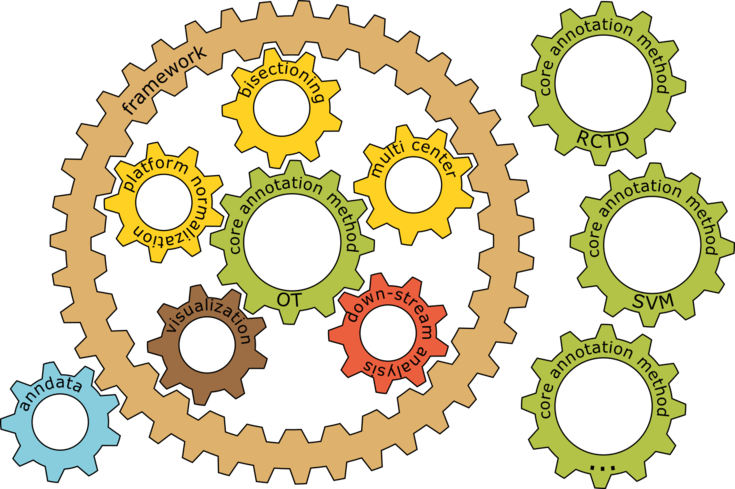
Rapid advances in single-cell-, spatial-, and multi-omics, allow us to profile cellular ecosystems in tissues at unprecedented resolution, scale, and depth. However, both technical limitations, such as low spatial resolution and biological variations, such as continuous spectra of cell states, often render these data imperfect representations of cellular systems, best captured as continuous mixtures over cells or molecules. Based on this conceptual insight, the authors build a versatile framework, TACCO (Transfer of Annotations to Cells and their COmbinations) that extends an Optimal Transport-based core by different wrappers or boosters to annotate a wide variety of data. They apply TACCO to identify cell types and states, decipher spatio-molecular tissue structure at the cell and molecular level, and resolve differentiation trajectories. TACCO excels in speed, scalability, and adaptability, while successfully outperforming benchmarks across diverse synthetic and biological datasets. Along with highly optimized visualization and analysis functions, TACCO forms a comprehensive integrated framework for studies of high-dimensional, high-resolution biology.
Original publication:
TACCO: Unified annotation transfer and decomposition of cell identities for single-cell and spatial omics.
Simon Mages, Noa Moriel, Inbal Avraham-Davidi, Evan Murray, Fei Chen, Orit Rozenblatt-Rosen, Johanna Klughammer, Aviv Regev, Mor Nitzan
Nature Biotechnology, 2023 Feb16 https://doi.org/10.1038/s41587-023-01657-3

