Halic Lab - Research
- Research
- Heterochromatin Formation
Initiation of heterochromatin formation
We used biochemical and high-throughput sequencing approaches to examine the mechanisms that mediate small RNA generation from the fission yeast centromeric repeats. Small RNA profiles in heterochromatin mutants demonstrated that generation of a subclass of siRNAs occurred independently of H3K9 methylation and HP1 protein. This immediately suggests that heterochromatin is not essential for siRNA generation. In contrary, siRNA generation precedes and guides heterochromatin formation in fission yeast. Furthermore, small RNA profiles show that generation of first siRNAs requires activities of RDRC, Dicer and Argonaute. In the nascent transcript cycle, siRNAs guide Argonaute to centromeric transcripts and recruit RDRC/Dicer to synthesize additional siRNAs. Our data show that initiation of siRNA generation has similar requirements as siRNA amplification. Since generation of first siRNAs also requires the slicer activity of Argonaute, this suggests that Argonaute-associated small RNAs guide RDRC/Dicer complex to centromeric transcripts to synthesize first dsRNA and first siRNAs. Although specific structural features in centromeric RNAs are likely to be important in initiation of silencing, requirement for the slicer activity of Argonaute for siRNAs generation suggests that Argonaute has to be loaded with a small RNAs prior to the generation of first siRNAs. This also suggests that another class of small RNAs has to exist in fission yeast.
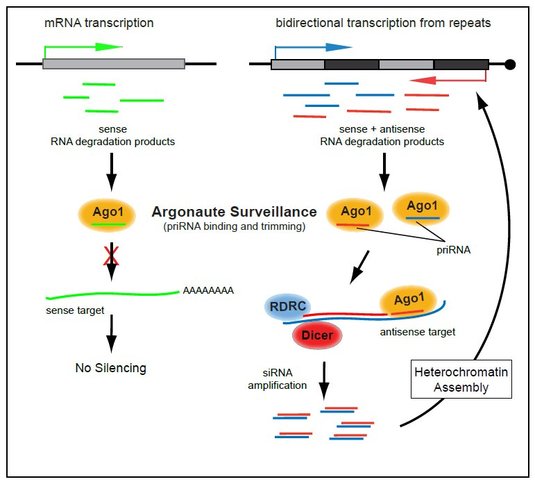 We have uncovered a distinct class of small RNAs, called primal small RNAs (priRNAs), which are generated independently of Dicer or RDRC and can guide Argonaute to specific transcripts to initiate silencing and heterochromatin formation. priRNAs originate from many abundant cellular RNAs, however, siRNA amplification and heterochromatin formation are limited to pericentromeric repeats. The basis of this specificity lies partly in the prevalence of bidirectional transcription within centromeric repeats, which generates both sense and antisense transcripts. Whereas, priRNAs cannot target most genomic regions containing protein coding genes and non-repetitive sequences which are sense to priRNAs, they can readily target repetitive regions that are bidirectionally transcribed because priRNAs will have antisense targets within these regions (see Figure). At the centromeric repeats, priRNAs can mediate low levels of H3K9 methylation in Argonaute dependent and Dicer independent way. Once priRNAs initiate siRNA generation at centromeric repeats and heterochromatin-independent siRNAs accumulate, RNAi machinery will target centromeric transcripts to induce H3K9 methylation. High levels of H3K9 methylation and siRNAs are then likely to nucleate the spreading of heterochromatin to induce transcriptional silencing at centromeric repeats.
We have uncovered a distinct class of small RNAs, called primal small RNAs (priRNAs), which are generated independently of Dicer or RDRC and can guide Argonaute to specific transcripts to initiate silencing and heterochromatin formation. priRNAs originate from many abundant cellular RNAs, however, siRNA amplification and heterochromatin formation are limited to pericentromeric repeats. The basis of this specificity lies partly in the prevalence of bidirectional transcription within centromeric repeats, which generates both sense and antisense transcripts. Whereas, priRNAs cannot target most genomic regions containing protein coding genes and non-repetitive sequences which are sense to priRNAs, they can readily target repetitive regions that are bidirectionally transcribed because priRNAs will have antisense targets within these regions (see Figure). At the centromeric repeats, priRNAs can mediate low levels of H3K9 methylation in Argonaute dependent and Dicer independent way. Once priRNAs initiate siRNA generation at centromeric repeats and heterochromatin-independent siRNAs accumulate, RNAi machinery will target centromeric transcripts to induce H3K9 methylation. High levels of H3K9 methylation and siRNAs are then likely to nucleate the spreading of heterochromatin to induce transcriptional silencing at centromeric repeats.
Our results suggest that a transcriptome surveillance mechanism based on the random association of small RNAs with Argonaute triggers RNAi-mediated heterochromatin formation within DNA repeats (see Figure). In this regard, the surveillance activity of Argonaute is reminiscent of the major histocompatibility antigen receptors, which act in proteome surveillance by binding and presenting protein degradation products. Although our results suggest that priRNAs are general RNA degradation products, it remains unclear how priRNAs are generated.
Our current work is focused on molecular mechanisms of small RNA mediated heterochromatin formation in fission yeast. We combine functional studies on the systemic level with genetic, biochemical and structural approaches, to work towards a mechanistic understanding of gene regulation in eukaryotic cells.

