Fröhlich Lab - Proteomics
- Fröhlich
- Group Members
- Publications
- Translational Proteomics
- Microplastics
- Embryo Proteomics
- News
Mammalian embryo and germ cell proteomics
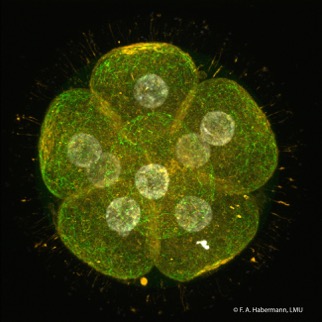 Mammalian reproduction is a complex process involving many important physiological and biochemical events, e.g., generation of mature gametes, fertilization, zygotic gene activation and embryonic development. Mammalian oocytes represent a highly specialized cell type since these cells contain the complete set of maternal transcripts and proteins essential for these processes, many of them synthesized and accumulated during oogenesis.
Mammalian reproduction is a complex process involving many important physiological and biochemical events, e.g., generation of mature gametes, fertilization, zygotic gene activation and embryonic development. Mammalian oocytes represent a highly specialized cell type since these cells contain the complete set of maternal transcripts and proteins essential for these processes, many of them synthesized and accumulated during oogenesis.
A subset of maternal proteins in the oocyte, referred to as „maternal effect proteins“ e.g., Nlrp5 or Npm2, play crucial roles during upcoming events, e.g., fertilization and embryogenesis.
While oocytes contain the complete set of molecular factors that can confer totipotency or pluripotency to somatic cells, and represent totipotent cells after fertilization, mature oocytes (MII stage) are unable to propagate on their own, and are condemned to cell death unless fertilization occurs.
In zygotes and embryos at initial cleavage cycles, transcription is silenced close to completion, and de novo translation is significantly reduced. As a consequence, macromolecular processes in early embryos occur almost exclusively at the level of proteins, making proteomic approaches particularly attractive for the analysis of oocyte and early embryonic development. As a model organism for these studies, we use the mouse and the bovine system. The latter shows strong similarities in estrus cycle (non-seasonal polyestric), singleton pregnancy and kinetics of early embryo development, thus reflecting crucial aspects of human reproductive biology. In addition, bovine oocytes are readily available, and effective protocols for in vitro fertilization (IVF) and in vitro production (IVP) of embryos to the blastocyst stage are well established.
Funded by the DFG research unit “Mechanisms of embryo-maternal communication”, we have analysed embryo-induced proteome changes in bovine endometrium (Berendt et al., 2005) and characterised dynamic alterations on the proteome level during in vitro maturation of bovine oocytes (Berendt et al., 2009).
On the oocyte level, we are currently studying the effect of donor age on the protein profiles in oocytes matured in vitro as well as in vivo, and characterise the oocyte secretome to identify proteins relevant for oocyte signalling. On the zygote and early embryo level, we analyse - on the resolution level of individual proteins - the onset of protein synthesis during embryogenesis and the impact of a delayed or impaired onset of protein synthesis. We recently began a molecular characterisation of relevant stages in early embryo development (zygote, 2-cell stage, 4-cell stage, morula, blastocyst) based on the expression kinetic and concentration of proteins selected for their involvement in developmentally relevant pathways.
For the establishment of these molecular signatures, the highly specific “Selected Reaction Monitoring” (SRM) is used. Current sensitivity of our multiplexed SRM approaches facilitates protein quantification in the attomole range from the protein equivalent of only a single oocyte or embryo. By determining absolute protein concentrations (i.e., molecules per embryo or blastomere), these data represent biochemical and biomedical parameters indispensable for the quantitative modelling and understanding of underlying biochemical networks.

