Wolf Lab - Research
- Research
- Monogenic Kidney Diseases
- Reproductive Biology
- Translational Animal Models
- Xenotransplantation
Early development and reproductive biology of livestock species
Maternal-to-embryonic transition is the period when maternal RNAs and proteins stored in the oocyte are gradually degraded and transcription of the embryonic genome is activated. The onset of embryonic transcription is obscured by the presence of maternal transcripts and could only be determined for genes which are not expressed in oocytes.
Using RNA sequencing (collaboration with Helmut Blum, LAFUGA) of bovine germinal vesicle and metaphase II oocytes, and of four-cell, eight-cell, 16-cell and blastocyst stage embryos, we established a comprehensive transcriptome data set of bovine oocyte maturation and early development.
The embryos investigated were produced by in vitro fertilization of German Simmental (Bos taurus taurus) oocytes with sperm from a single bull of the genetically distant Brahman (Bos taurus indicus) breed to obtain a large number of single nucleotide polymorphisms (SNPs) for identification of the parental origin of transcripts. Further, RNA-Seq libraries were produced without polyA+ selection enabling the identification of intronic sequences in transcripts, which can be found in de novo synthetized transcripts due to incomplete co-transcriptional splicing and can thus be used to discriminate them from spliced maternal transcripts stored in the oocyte. EGA was analyzed by i) detection of embryonic transcripts which are not present in oocytes; ii) detection of transcripts from the paternal allele; and iii) detection of primary transcripts with intronic sequences.
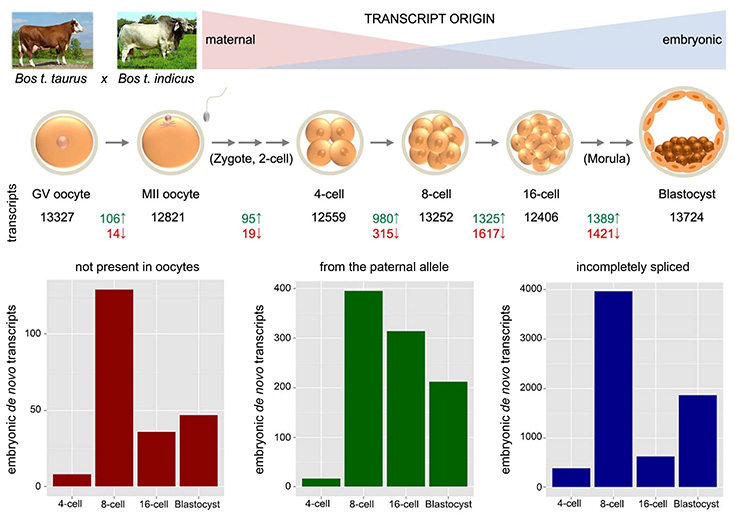
Mapping of gene activation in early bovine embryos.
Using these approaches we were able to map the onset of embryonic transcription for almost 7,400 genes (Graf et al., PNAS 111: 4139-44, 2014). RNA-Seq analyses provided new insights into embryonic genome activation, allelic expression of genes, and transcriptional networks governing early development of embryos. RNA-Seq data provide a fundamental basis for embryo biosystems research, including germ cell potential and genetic, epigenetic and environmental disturbances of early development. In addition, studies based on structured illumination microscopy revealed major changes in higher order chromatin organization associated with major EGA (Popken et al., Nucleus 5: 555-89, 2014).
Current studies address EGA in cloned embryos as well as developmental heterogeneity and early lineage specification events in bovine embryos at the time of major EGA. In addition, gene editing (CRISPR/Cas9) is used to dissect the roles of pluripotency networks in early bovine development.

