Jae Lab - Research
- Research
- Methods
Functional Genomics
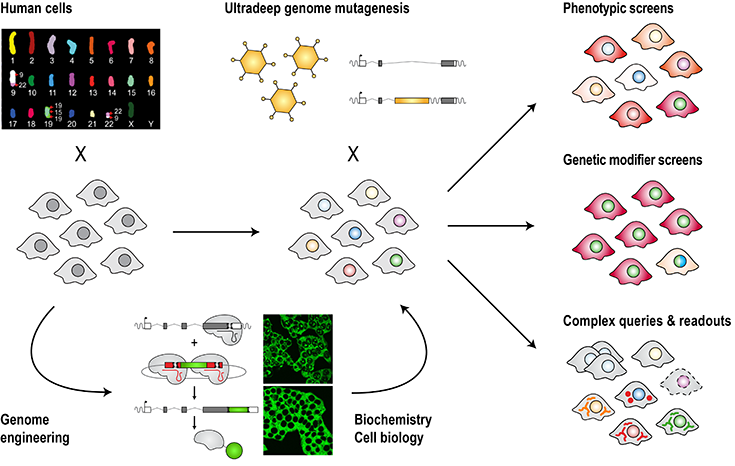
Virtually every form of human disease is shaped by the individual or concerted action of genes. Incredibly successful work in model organisms such as yeast has paved the way to build genetic wiring diagrams of the eukaryotic cell. However, strategies for comparable experiments in a human system have long been hindered by the diploid nature of the human genome, which effectively buffers against individual genetic alterations.
To gain unbiased insights into the inner workings of human cells, we combine CRISPR-mediated genome engineering and synthetic biology with ultradeep mutagenesis and phenotypic screening in the human system. This has led to a suite of discoveries across different areas of biology ranging from signal transduction, to infection and immunity, to organelle function (e.g. Fessler et al., Nature, 2020; Brockmann et al., Nature, 2017; Mezzadra et al., Nature, 2017; Blomen et al., Science, 2015; Jae et al., Science, 2014; Jae et al., Science, 2013).
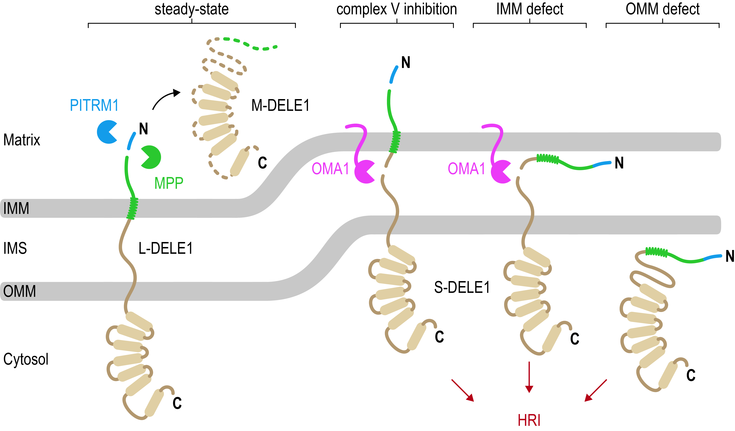
Mitochondrial fidelity
Mitochondria fulfill a wide range of important cellular functions, constantly experience stress, and malfunction in aging and severe human diseases, such as different forms of neurodegeneration, as well cardiovascular defects and cancer. Surprisingly, mitochondria do not encode any stress response genes of their own and instead need to alert the cell about threats to their integrity and function. We are only beginning to understand the complex cross-talk that underlies organelle homeostasis in the human system and unbiased genome-wide experiments are urgently needed. Using our functional genomics platform, we recently discovered the pathway that signals mitochondrial stress to the cytosol in human cells: the previously little-studied mitoprotein DELE1 is cleaved by stress-activated mitoprotease OMA1, which results in the redistribution of the C-terminal cleavage fragment (S-DELE1) to the cytosol, where it binds to and activates a kinase called HRI. In turn, the cell attenuates normal translation of most mRNAs in favor of a few select messages encoding the information necessary to restore cellular protein homeostasis and mitochondrial health (Fessler et al., Nature, 2020; Eckl et al., CMLS, 2021; Fessler et al., Nature Commun., 2022). This discovery opens up a new field in mitochondria research with key questions that need to be answered.
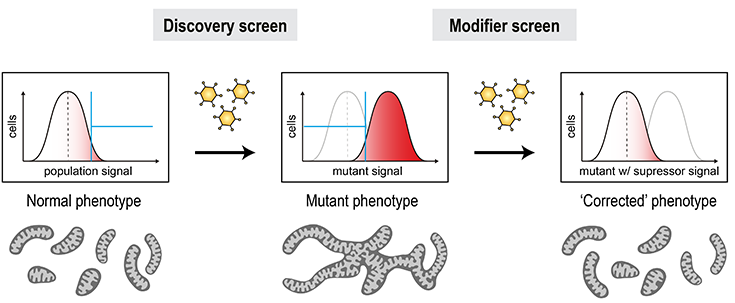
Genetic suppression of organelle defects
Mitochondrial defects represent the largest group of human inborn errors with unmet clinical need in many cases. Genetic organelle defects typically result from loss-of-function mutations, which render the condition ‘undruggable’ by classical means. However, the identification of genetic interactions of disease alleles could open new possibilities to restore cellular function through indirect targeting of genetic suppressors. Mitofusins play important roles in shaping of the mitochondrial network, contacts with other organelles, and mitochondrial removal by mitophagy. Their activity thus needs to be delicately balanced to meet the respective requirements of cells in different settings. Defects in these processes can result in conditions like CMT and PD. With our unique pipeline, it is possible to engineer patient defects at the cellular level and systematically scan the genome for genetic suppressors that restore a functional state.