Herzog Lab - Research
- Research
- Integrative Structural Biology
Integrative Structural Biology
The XL-MS approach is especially valuable for protein assemblies that cannot be purified as stable homogeneous particles and are refractory to established structural biology techniques. The length of the bi-functional agent defines a critical distance restraint (Cα-Cα ~ 27 Å) for integrative structural biology studies. Work on RNA polymerases revealed that 95% of inter-protein crosslinks indicate physical contacts between proteins.
In a collaborative effort, we have recently studied the architecture of the budding yeast chromatin remodeler, INO80, interacting with its nucleosome substrate by combining XL-MS and cryo-electron microscopy. Although X-ray structures for only four of the 15 subunits have been available, about 200 inter-protein crosslinks indicate the spatial proximity of protein domains. The functional interpretation at 'motif resolution' has indicated how INO80 subunits recognize the nucleosome substrate and facilitate histone exchange. This study demonstrates that hybrid structural analyses using XL-MS derived distance restraints provide detailed mechanistic insights even in the absence of high resolution structural information and establishes XL-MS as a key technology for the topological analysis of native chromatin-assembled protein complexes.
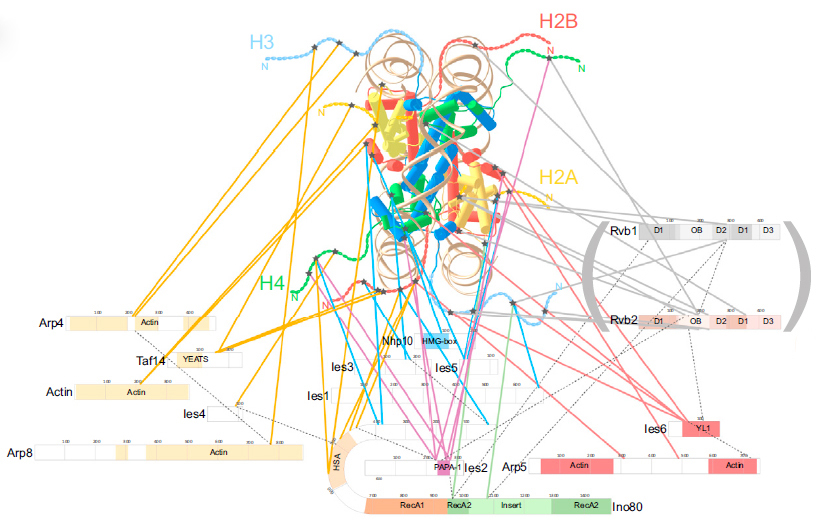
Integrative structural model of the INO80 chromatin remodeler in complex with its nucleosome subtrate using crosslink-derived distance restraints (Tosi et al., 2013).

