Herzog Lab - Research
- Research
- Integrative Structural Biology
Interaction proteomics
Cellular processes can rarely be attributed to the activity of a single protein. Instead, proteins act in functional modules, such as macromolecular complexes or signal transduction networks. Gaining mechanistic insights into the function of multi-subunit modules frequently involves their structural elucidation.
Chemical crosslinking, in combination with mass spectrometry (XL-MS), is a low-resolution structural technique for the characterization of the architecture of proteins and their domains in a complex. XL-MS provides distance information by identifying proximate lysine residues that have become covalently linked in a protein complex with a bi-functional crosslinking reagent. This approach has been successfully applied to protein complexes purified for crystallographic purposes.
In contrast to high resolution structural biology techniques, XL-MS has the potential to acquire spatial restraints from heterogeneous and less concentrated protein samples. This method is thus unrivalled in its capability to characterize the topology of signaling pathways or cellular compartments by systematic analysis of endogenous protein complexes.
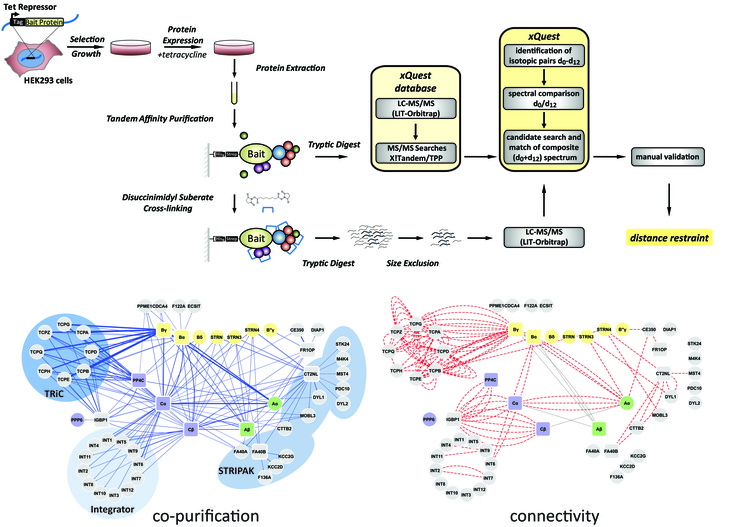
Schematic workflow of chemical crosslinking and mass spectrometry of affinity-purified protein complexes in a protein phosphatase 2A signalling network (Herzog et al., 2012).

