Klughammer Lab - Research
- Research
- Projects
Deciphering complex cellular systems based on their native high-dimensional representations
In multicellular organisms, it is essential that cells work together in concerted ways while fulfilling dedicated functions to keep the organism as a whole well and healthy despite an abundance of internal and external challenges. These astonishingly robust systems are enabled through self-regulatory molecular circuits within and across cells that evolved over millions of years. Nevertheless, sometimes things can go wrong, leading to disease or even death.
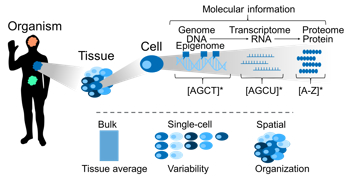
Conveniently, a cell’s functionality is largely defined by its molecular setup in the form of DNA, RNA, and protein – information bearing sequences that we can efficiently read using high-throughput methods. Measured in bulk, at single-cell resolution or spatially resolved, the resulting high-dimensional data allow us to characterize and model cellular types, states, programs, and interactions in diverse biomedical contexts ranging from host-pathogen interactions [3], over malignant diseases [2] and charting complex tissues [1,4], to evolutionary questions [5]. Our research is primarily data-driven while combining wet-lab and computational methods. To efficiently leverage large amounts of high-dimensional data, we use and develop computational approaches including data-processing and integration frameworks, machine learning, and visualizations in a high-performance computing environment.
1. High-definition spatial transcriptomics for in situ tissue profiling.
Vickovic S, Eraslan G*, Salmén F*, Klughammer J*, Stenbeck L*, Schapiro D, Äijö T, Bonneau R, Bergenstråhle L, Navarro JF, Gould J, Griffin GK, Borg Å, Ronaghi M, Frisén J, Lundeberg J, Regev A, Ståhl PL. Nat Methods. 2019 Oct;16(10):987-990. PubMed
2. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space.
Klughammer J*, Kiesel B*, Roetzer T, Fortelny N, Nemc A et al. Nat Med. 2018 Oct;24(10):1611-1624. PubMed
3. Comparative Genome Sequencing Reveals Within-Host Genetic Changes in Neisseria meningitidis during Invasive Disease.
Klughammer J, Dittrich M, Blom J, Mitesser V, Vogel U, Frosch M, Goesmann A, Müller T, Schoen C. PLoS One. 2017 Jan 12;12(1):e0169892. PubMed
4. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types.
Li J*, Klughammer J*, Farlik M*, Penz T*, Spittler A, Barbieux C, Berishvili E, Bock C, Kubicek S. EMBO Rep. 2016 Feb;17(2):178-87. PubMed
5. Differential DNA Methylation Analysis without a Reference Genome.
Klughammer J, Datlinger P, Printz D, Sheffield NC, Farlik M, Hadler J, Fritsch G, Bock C. Cell Rep. 2015 Dec 22;13(11):2621-2633. PubMed

