Hopfner Lab - Research
- Research
- Chromatin Remodeller
- Innate Immunity
- Therapeutic Protein Engineering
- OPSYON Spin-off Project
Chromatin Remodeller
DNA resides in the eukaryotic cell's nucleus as an assembly with myriads of proteins, collectively referred to as chromatin. The major and basic structural units of chromatin are nucleosomes, ~146 base pairs of DNA tightly wrapped around a histone protein octamer. Nucleosomes package and protect the DNA, but also carry positional and chemical information and play important functions in gene expression, genome maintenance and in specifying cell identity.
The position and composition of nucleosomes along chromosomes is not random and a defined nucleosomal landscape is created and maintained by the collective activity of chromatin remodeller and chromatin modifying factors. Chromatin remodellers are intriguing molecular machines that use the free energy of ATP hydrolysis to position, slide or edit nucleosomes, in order to properly structure chromatin and to regulate and facilitate chromosomal transcription, replication and repair.
The lab pioneered structural studies of remodellers, starting from X-ray structure of the conserved and family defining Swi2/Snf2 ATPase domain - the DNA groove tracking "motor" of remodeller - in complex with DNA to a recent high resolution cryo-EM structure or the core of the 19 subunit megadalton remodeller INO80 complex bound to the nucleosome.
Future studies will aim at determining structures of full remodelling complexes in functional states in order to derive a detailed structural and mechanistic understanding of how these molecular machines can precisely position and edit nucleosomes. A long term-goal is to understand at the structural level the interplay of remodelling molecular machines and other chromosomal factors in shaping chromatin and how these proteins are (mis)regulated in health and disease.
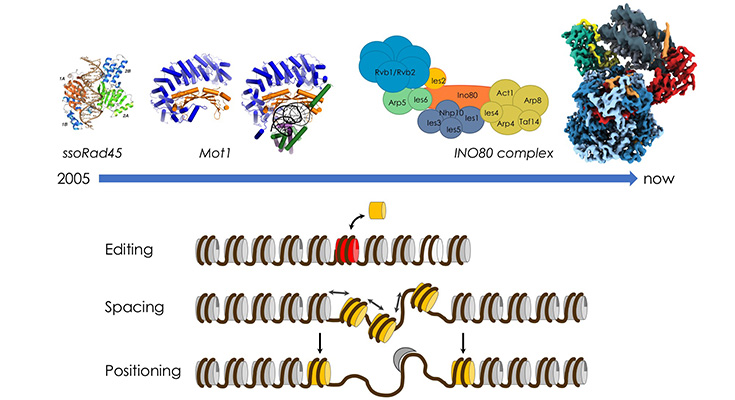
Figure: Remodeller Structures determined by our group: ssoRad45, Mot1 and the INO80 complex.
Publications:
Structural basis for ATP dependent chromatin remodelling by the INO80 complex.
Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner KP.
Nature. 2018 April 11; doi:10.1038/s41586-018-0029-y. PubMed
Structural basis for recognition and remodeling of the TBP:DNA:NC2 complex by Mot1.
Butryn A, Schuller JM, Stoehr G, Runge-Wollmann P, Förster F, Auble DT, Hopfner KP.
Elife. 2015 Aug 10;4. doi: 10.7554/eLife.07432. PubMed
Structure and Subunit Topology of the INO80 Chromatin Remodeler and Its Nucleosome Complex
Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner KP.
Cell, Volume 154, Issue 6, 1207-1219, 12 September 2013. PubMed
Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Wollmann P, Cui S, Viswanathan R, Berninghausen O, Wells MN, Moldt M, Witte G, Butryn A, Wendler P, Beckmann R, Auble DT, Hopfner KP.
Nature. 2011 Jul 6;475(7356):403-7. doi: 10.1038/nature10215. PubMed
X-Ray Structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase Core and Its Complex with DNA.
Dürr H, Körner C, Müller M, Hickmann V, Hopfner KP.
Cell. 2005 May 6;121(3):363-73. PubMed
Reviews:
Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines.
Hopfner KP, Gerhold CB, Lakomek K, Wollmann P.
Curr Opin Struct Biol. 2012 Apr;22(2):225-33. Epub 2012 Mar 23. PubMed

