Hopfner Lab - Research
Innate Immune RNA and DNA Sensing
Cytosolic DNA and viral RNA are powerful trigger of the innate immune system, which is a first line of defense against microbial pathogens. Specialized receptors detect microbial or danger associated molecular patterns and induce cellular responses by changing the cell's gene expression profile, triggering inflammatory reactions and activating the adaptive immune system.
Our lab investigates the molecular and structural mechanism of the detection of nucleic acids and other molecular patterns by innate immune receptors and how receptor activation triggers downstream signaling cascades. Past and recent achievements include the structural mechanism of RIG-I and cGAS nucleic acid sensors and the discovery of the bacterial second-messenger c-di-AMP. Our research combines structural methods (X-ray crystallography and cryo-EM) with a variety biophysical, biochemical and cell-based assays.
A particular focus of our current and future research is to understand qualitatively and quantitatively how the innate immune system distinguishes self from non-self. Under certain conditions, for instance through inherited or sporadic gain of function mutations in the receptors or through induction of cellular stress, activation of innate immune receptors can lead to recognition of self nucleic acids, sterile inflammation and autoimmune diseases. Underlying structural and molecular mechanisms aid the development of innate immune modulators in therapeutic approaches.
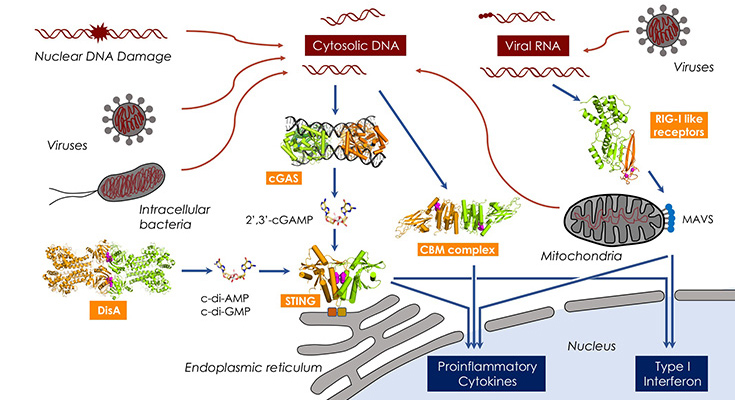
Figure: Detection of nucleic acids and other molecular patterns by innate immune receptors and receptor activation triggers downstream signaling cascades. Structural studies on DisA, cGAS, STING, the CBM complex and RIG-I-like receptors.
Publications:
cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders.
Andreeva L, Hiller B, Kostrewa D, Lässig C, de Oliveira Mann CC, Jan Drexler D, Maiser A, Gaidt M, Leonhardt H, Hornung V, Hopfner KP.
Nature. 2017 Sep 21;549(7672):394-398. doi: 10.1038/nature23890. Epub 2017 Sep 13. PubMed
ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA.
Lässig C, Matheisl S, Sparrer KM, de Oliveira Mann CC, Moldt M, Patel JR, Goldeck M, Hartmann G, García-Sastre A, Hornung V, Conzelmann KK, Beckmann R, Hopfner KP.
Elife. 2015 Nov 26;4. pii: e10859. PubMed
Structural mechanism of cytosolic DNA sensing by cGAS.
Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP.
Nature. 2013 May 30. doi: 10.1038/nature12305 PubMed
Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates.
Witte G, Hartung S, Büttner K, Hopfner KP.
Mol Cell. 2008 Apr 25;30(2):167-178 PubMed
Reviews:
OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids.
Hornung V, Hartmann R, Ablasser A, Hopfner KP.
Nat Rev Immunol. 2014 Aug;14(8):521-8. doi: 10.1038/nri3719. Epub 2014 Jul 18. PubMed

