Hopfner Lab - Research
Genome Maintenance
The integrity of DNA and the information contained in genomes is under constant threat by extrinsic factors, for instance UV-light and genotoxic chemicals, but also intrinsically through mutations and DNA breaks generated during e.g. DNA replication. A vast number of DNA lesions are experienced by a human cell per day, and the resulting damage can lead to cell death, senescence and cancer.
DNA double-strand breaks (DSBs) are one of the most problematic DNA lesions. In contrast to other types of lesions, where the damaged strand of DNA is repaired using the non-damaged strand as template, DSBs affect both strands and disrupt the flow of the genetic information. DSBs often cause tumorigenic chromosomal aberrations, as for instance seen in the increased cancer susceptibility caused by mutations in the DNA double-strand repair proteins BRCA1, BRCA2, NBS1 and ATM.
To deal with DNA double-strand breaks, eukaryotic cells possess a complex DNA damage response (DDR), which halts the cell cycle, remodels and structures chromatin, invokes DSB repair pathways or induces programmed cell death. We study the mechanism of DNA double-strand break repair using a combination of structural and functional studies. A key goal is to understand how DNA ends are detected and processed by the Mre11-Rad50-Nbs1 complex and its interacting repair and chromatin factors. Our main tool is cryo-EM, but we also employ X-ray crystallography, small angle X-ray scattering and mass spectrometry, complemented by biochemical and cell-based studies. Long term-goals are revealing the structural-molecular choreography of DNA double-strand break repair in chromatin and its molecular pathology in cancer and other human diseases.
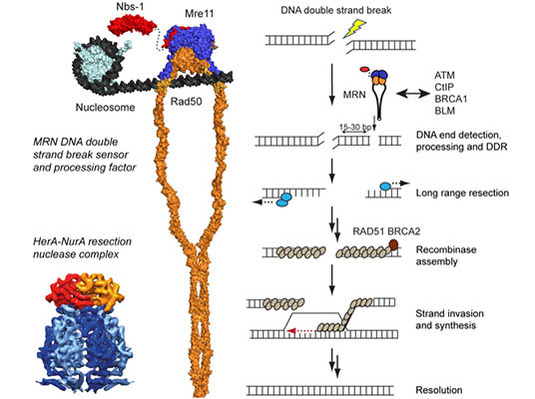
Figure: Model of the Mre11-Rad50-Nbs1 (MRN) complex bound to a nucleosome containing DNA, along with a cryo-EM structure of the archaeal HerA-NurA DNA resection complex and a general scheme of homologous recombination.
Publications:
Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner KP.
EMBO J. 2016 Apr 35(7):759-72. PubMed
Structure of the Rad50 DNA double-strand break repair protein in complex with DNA.
Rojowska A, Lammens K, Seifert FU, Direnberger C, Feldmann H, Hopfner KP.
EMBO J. 2014 Oct 33(23):2847-59. PubMed
Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Möckel C, Schele A, Strässer K, Jackson SP, Hopfner KP.
Nat Struct Mol Biol. 2012 Jun 19(7):693-700. PubMed
The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair.
Lammens K, Bemeleit DJ, Möckel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermüller C, Söding J, Strässer K, Hopfner KP.
Cell. 2011 Apr 145(1):54-66. PubMed
Reviews:
Invited review: Architectures and mechanisms of ATP binding cassette proteins.
Hopfner KP.
Biopolymers. 2016 Mar 105(8):492-504. PubMed

