Stigler Lab - Research
Establishment of Hierarchically Structured Chromosomes
Interphase chromosomes in higher eukaryotes are hierarchically structured on different levels, ranging from DNA wrapped around histones, to chromatin fibers, up to large chromosomal domains encompassing hundreds of kilobases. Hi-C experiments have shown that these topologically associating domains (TADs), which consist of regions of increased interactions of certain genomic regions, are a structural feature of all higher eukaryotes. TADs are generally isolated by domain boundaries from adjacent regions.
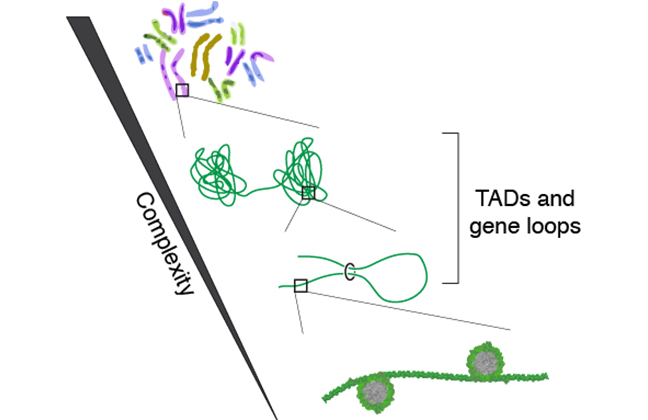 Figure: Chromosomes are organized hierarchically on different length scales.
Figure: Chromosomes are organized hierarchically on different length scales.
Current models claim that TADs are formed by the action of chromosomal architectural proteins, chiefly members of the SMC (structural maintenance of chromosomes) family, such as cohesin or condensin. With the help of genomic insulators, such as CTCF, SMC proteins act as intra-molecular crosslinkers and tether distant genomic sites together. The thus-established three-dimensional architecture of chromosomes is directly linked to gene regulation.
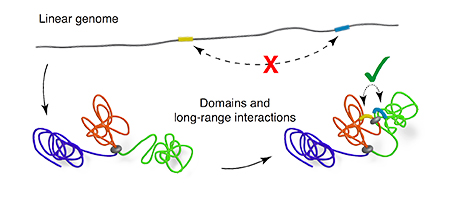 Figure: Chromosomal binding proteins, such as SMC proteins and CTCF, are the drivers of chromosomal rearrangements into compartments. Genomic elements that are distant on a linear genome can come into close contact in the folded configuration.
Figure: Chromosomal binding proteins, such as SMC proteins and CTCF, are the drivers of chromosomal rearrangements into compartments. Genomic elements that are distant on a linear genome can come into close contact in the folded configuration.
The three-dimensional structure of chromosomes ensures that enhancer and repressor elements become spatially close to promoters that, on a linear genome, may be hundreds of kilobases away.
Our lab has set out to decipher the protein-protein and protein-DNA interactions of chromosomal architectural proteins that lead to the formation of three-dimensionally folded chromosomes.

